Since 2011, we have focused on a variety of research topics and directions, such as protein structural modeling, gene ontology analysis, etc. Current research focus of our group involves computational analysis of disease pathomechanisms, telomere bioinformatics and regulation of alternative splicing. This page highlights the main research directions, omitting the old, as well as the very new ones.
Computational analysis of disease pathomechanisms
Pathway-centered analysis of high-throughput data
Cell signaling pathways are sets of directed interactions between biological molecules, that are initiated by a particular signal (e.g. a ligand binding to a receptor) and result in realization of certain target processes (e.g. transcription of genes).
We try to understand how the activity state of the pathways changes during disease development. In contrast to gene-centered analysis, this approach accounts for the interactions between gene products and provides biological significance of the observed disturbances. For this we use gene expression data and pathway topology information obtained from publicly available databases, such KEGG, and manually edited or curated ones. We than apply algorithms for pathway signal flow calculation.
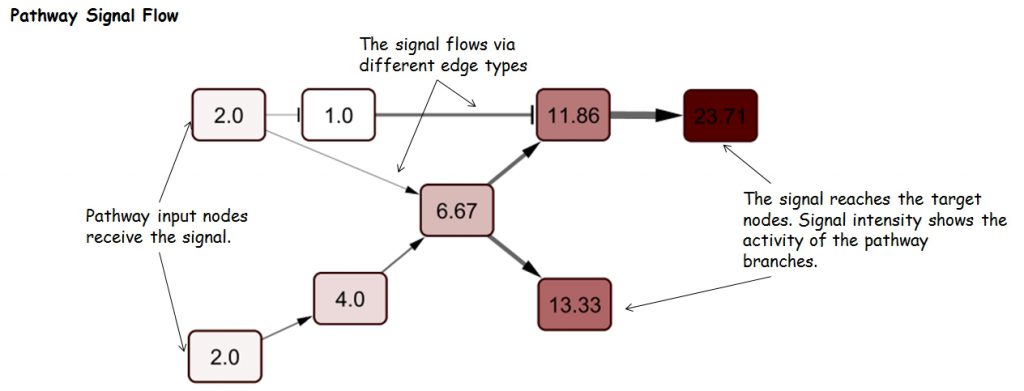
Pathway Signal Flow (PSF), or perturbation, is the flux generated by propagation of the signal starting from input nodes, flowing through intermediate nodes in branches and accumulating at sink nodes. Thus, PSF can be an indicator of pathway activity state. Assessment of changes in pathway activity is of major interest for identification of processes involved in the formation of certain phenotypes (healthy and diseased states), and assessment of cell response to drugs and other stimuli.
Software tools
We have developed software packages for R, Matlab and Cytoscape (see the Software list) to allow for pathway parsing, editing, tuning and applying PSF algorithms on pathway topologies.
 KEGGParser: a tool for parsing and editing KEGG pathway graphs in Matlab. Imports KEGG pathways into Matlab by creating a biograph object, and allows to edit nodes and edges and save the edited graphs for further analysis.
KEGGParser: a tool for parsing and editing KEGG pathway graphs in Matlab. Imports KEGG pathways into Matlab by creating a biograph object, and allows to edit nodes and edges and save the edited graphs for further analysis.
CyKE GGParser: a Cytoscape app for parsing, editing and tuninig of KEGG pathway maps. Downloads and imports KEGG pathways into Cytoscape. Applied automati corrections for missing nodes and edges. Provides functionality for tuning the pathways based on tissue-specific gene expression and protein-protein interactions.
GGParser: a Cytoscape app for parsing, editing and tuninig of KEGG pathway maps. Downloads and imports KEGG pathways into Cytoscape. Applied automati corrections for missing nodes and edges. Provides functionality for tuning the pathways based on tissue-specific gene expression and protein-protein interactions.
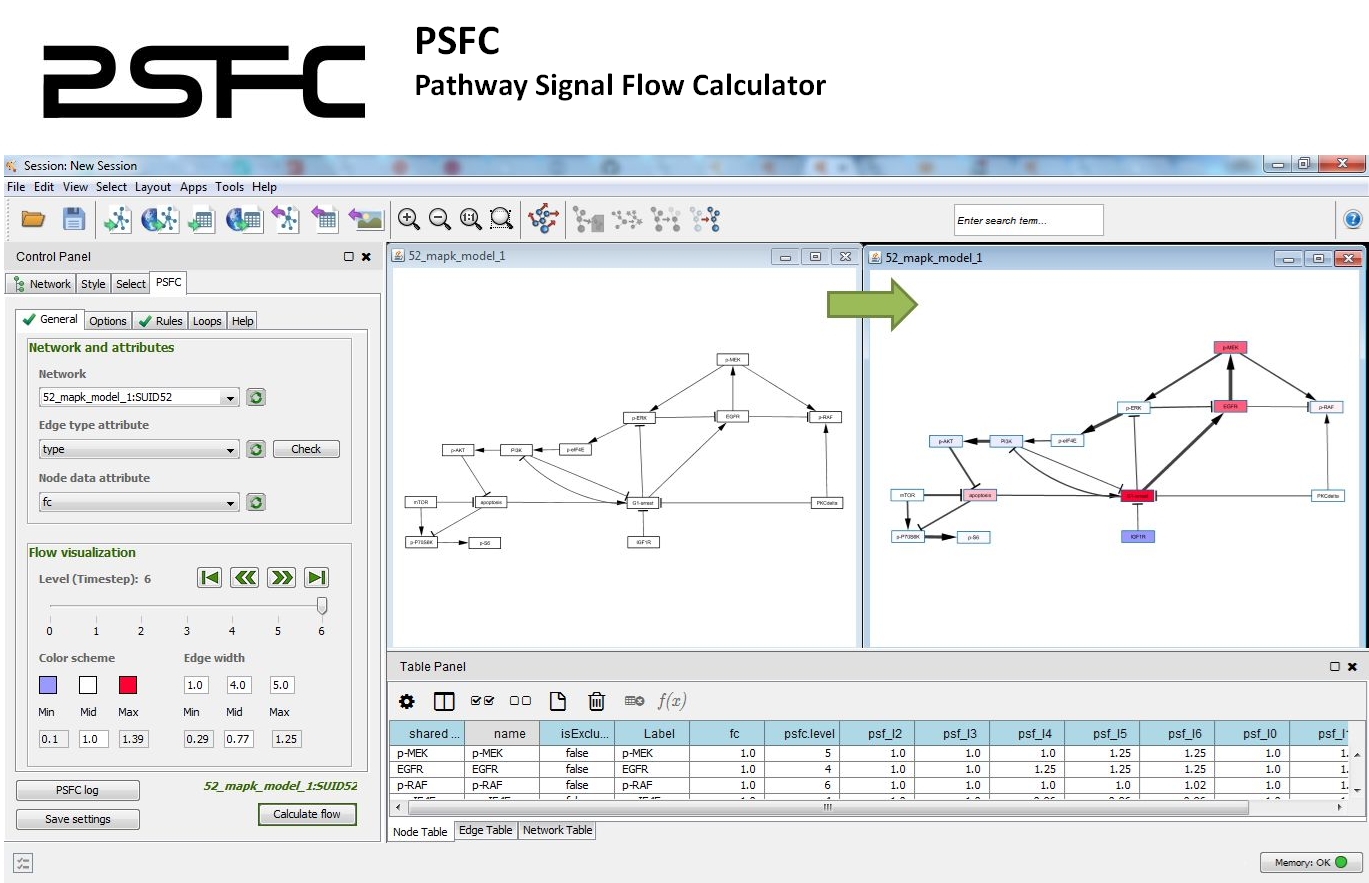 PSFC: Pathway signal flow calculator app for cytoscape.
PSFC: Pathway signal flow calculator app for cytoscape.
PSFC allows for using custom algorithms and mathematical functions to characterize the signal flow within a pathway graph.
The user may choose for:
– Mathematical funtions can to edges of different types for processing source to target signal transfer.
– Algorithms for processing multiple signals entering or leaving a node.
– Algorithms for handling of feedback loops.
– Signifcance calculation options.
The signal flow is visualized within the Cytoscape environment and a report file is generated for quantitative analysis.
Pathway deregulation landscapes in complex diseases
Complex human disea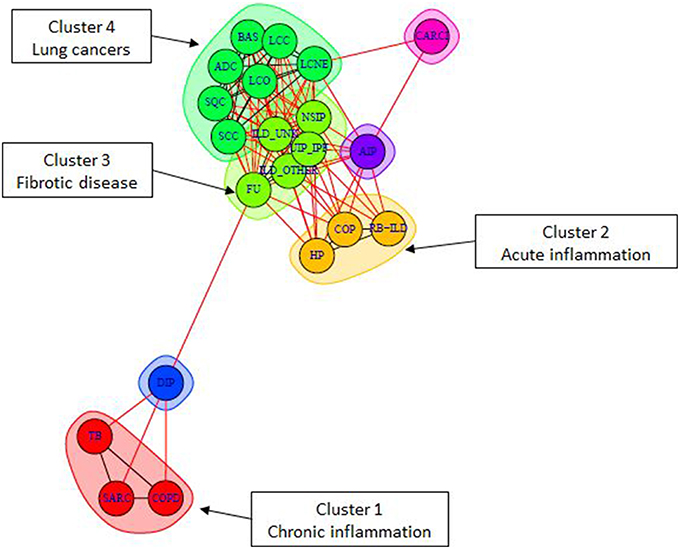 ses are often described concerted changes in various aspects of cellular life: from mutations to epigenetic changes and associated changes in transcriptome. Thus, causes and pathomechanisms of these diseases are often difficult to uniquely describe and classify. They are often classfied and described by common symptoms, e.g. chronic and acute lung diseases, autoimmune and autoinflammatory diseases, etc.
ses are often described concerted changes in various aspects of cellular life: from mutations to epigenetic changes and associated changes in transcriptome. Thus, causes and pathomechanisms of these diseases are often difficult to uniquely describe and classify. They are often classfied and described by common symptoms, e.g. chronic and acute lung diseases, autoimmune and autoinflammatory diseases, etc.
Our research aims at describing these diseases by pathway activity perturbations and classfying them according to commonly disregulated pathways, as well discovering disease-specific pathways. The objective of our approach is, on one hand, to better understand the global picture of disease pathomechanisms and their similarities and specificities, and, on the other hand, to provide theoretical bases for developing drugs and treatments for groups of similar diseases.
Telomere bioinformatics
Telomere length calculation from NGS data
Telomere length homeostasis plays an important role in cell fate regulation, differentiation, ageing and disease development. However, little is known about the mechanisms of telomere length regulation and the processes that depend on telomere length dynamics. In order to facilitate telomere biology research via utilization of next generation sequencing technologies, we have developed a program Computel to measure the mean length of telomeres from Illumina Whole Genome Sequencing data. Computel performs by alignment of NGS reads to a specially designed telomeric index. It is robust to sequencing errors and is adjusted to avoid capturing false positive reads, such as those from interstitial telomeric regions in the genome.
We have used Computel in a quantitative trait association study for the South Asian genomes. There are several ongoing projects, where data from several cancer types are being analyzed with Computel.
Analysis of telomere length regulation
Telomere length is dependent on two counteracting processes: environmental and genetic factors leading to telomere shortening and telomere elongation processes. Tha latters are of two types: elongation of telomeres by a nucleoprotein complex telomerase and alternative mechanisms of telomere elongation (ALT) that depend on homologous recombination events. Stem cells and the majority of cancer cells elongate their telomeres via expression of telomerase, whereas 10-15% of cancer cells utilize ALT. To make the telomerase-therapies more efficient and for better diagnostic and prognostic tests, it’s important to identify the active telomere elongation mechanism.
We are engineering biomolecular pathways involved in telomerase-dependent and alternative lengthening mechanisms of telomeres and computational approaches for validation of these pathways. Our final aim is computational identification of the telomere regulation state of the cell, based on transcriptome, genome, and epigenome data.
Association of telomere length dynamics with transcriptome and epigenome
Telomere length dynamics is regulated by complex interplay between telomere elongation and shortening processes. Several genes are involved in regulation of telomere length in healthy and diseased conditions. While many of the gene players have already been identified, there is still a gap to fill in this area. Changes in gene expression caused by genetic variations and epigenetic modifications, may lead to elongation or shortening of telomeres.
On the other hand, telomere length dynamics may lead to variation of expression of certain genes. This regulation may be performed via a phenomenon called the telomere position effect (TPE), a reversible silencing of genes located near telomeres, caused by spreading of the telomeric heterochromatin to near by genomic regions; and by long distance effects caused by telomere looping and spacial rearrangements of chromosomes in the nucleaus (TPE over long distances).
In order to identify novel genes regulating or regulated by telomere length dynamics, we develop computational methods for identification of associative relationship between telomere length dynamics and respective changes transcriptomic and epigenomic profiles of genes.
Association of telomere length dyanamics with complex diseases and cancers
Telomere length regulation plays a crucial role in development of age-related diseases and cancers, since their gradual shortening through cell division cycles leads may lead to telomeric dysfunction, chromosomal instability and senescence. Additionally, gradual changes in telomere length may lead to variations in expression of genes, which may have a role in disease pathomechanisms.
We utilize our computational approaches for telomere length measurement and associative studies to investigate the role of telomeres in molecular mechanisms of disease progression.
SARS-CoV-2 bioinformatics
Genetic analysis of SARS-CoV-2 in Armenia
The study aims at genetic characterization of clinical isolates of SARS-COV-2 in Armenia using whole-genome nanopore sequencing. This study can contribute to public health actions by providing a phylogenetic structure of disease outbreak, trace transmission networks, identifying new mutations in the viral genome associated with changes in transmission rates, disease course, and interference with diagnostic tests.
Drug repositioning for COVID-19 with multimodal biological data
The study aims at development of pathway/network-based drug repositioning approach to effectively predict already approved drugs or their combinations for COVID-19 treatment. We use systematic knowledge about protein interactions of human and SARS-CoV-2 and -omics data. We apply artificial intelligence algorithms and network analysis tools to integrate collected data for effective predictions. An online platform will be developed for network-based investigation of repurposed drugs which will be widely accessible for clinicians and researchers.